Читать книгу Structure and Function of the Bacterial Genome - Charles J. Dorman - Страница 15
1.4 Chromosome Replication: Elongation
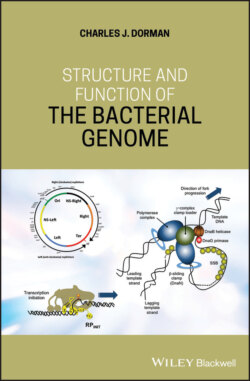
ОглавлениеOnce replication has been initiated, the replisome is responsible for progressive DNA synthesis during the elongation phase of chromosome replication. This large complex is composed of a pentameric clamp loader, the DNA polymerase clamp (DnaN), the three‐subunit DNA primase (DnaG), and the hexameric helicase DnaB (Bailey et al. 2007; Reyes‐Lamothe et al. 2010) (Figure 1.4). The helicase uses ATP hydrolysis to unwind the DNA duplex, moving along the lagging strand of the DNA as it does so. Single‐stranded DNA‐binding protein (SSB) coats the separated ssDNA strands, thus preventing reformation of the duplex by religation and attack by nucleases (Beattie and Reyes‐Lamothe 2015).
The primase, DnaG, possesses a central RNA polymerase domain where the RNA primers used in DNA synthesis are manufactured (Corn et al. 2008). The primer emerges from the DnaG‐DnaB complex and is transferred to DNA polymerase and SSB (Corn et al. 2008). DNA Polymerase III works with the beta‐clamp protein (DnaN) to extend the primer, creating a new DNA strand at a rate of 1000 bases per second (Beattie and Reyes‐Lamothe 2015). It is advantageous to have DnaN as a component of the replisome because a beta‐clamp must be reloaded for the synthesis of each lagging strand Okazaki fragment (Beattie and Reyes‐Lamothe 2015). If the replication fork stalls or breaks, replication can be restarted through a DnaA‐independent mechanism. Here, the PriA helicase, in association with accessory proteins such as PriB, PriC, and DnaT, binds to the gapped replication fork and loads DnaBC. In some cases, the restart may be associated with a strong transcription promoter that generates an R‐loop where PriA can introduce DnaBC on the displaced DNA strand (Heller and Marians 2006; Kogoma 1997). Of the approximately 300 copies of DNA gyrase that are bound to the E. coli chromosome at any one time, about 12 accompany each moving replication fork to manage the DNA topological disturbance that is associated with fork migration (Stracy et al. 2019).